|
Jmol: Open-source
molecular visualization and analysis |
|
Robert M.
Hanson (St. Olaf College), Egon
Willighagen (Cologne University Bioinformatics Center), Nicolas
Vervelle (Jmol Project), Timothy
Driscoll (molvisions), and Miguel Howard
(Jmol Project) |
Introduction to Jmol
|
Jmol is a free,
open-source molecule viewer for chemistry and biochemistry. It works on
multiple platforms, including Windows, Mac OS X, and Linux/Unix systems. The
software consists of three parts, all written in the Java programming
language: the Jmol applet, the Jmol application, and the Java development
toolkit, JmolViewer. The Jmol application is a standalone Java application
that runs on the desktop. JmolViewer is a set of Java “classes” that can be
integrated into other Java applications to provide molecular visualization
and analysis of chemical structure. While interesting in their own right, in
this paper we will focus on the third component of the triad, the Jmol
applet, as it is the component that can be integrated into web pages. |
|
|
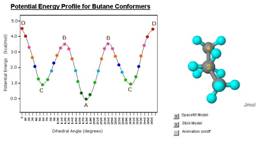
Jmol has many features to offer in the fields of chemistry and biochemistry. We will highlight many of them in this paper. Screen shots illustrating the broad variety of applications of Jmol are shown in Figure 1. For example, Jmol supports rendering of secondary structures of proteins and nucleic acids, it can produce animations of chemical reactions and conformational changes, and it can help visualize vibrations of small molecules based on quantum mechanics or molecular mechanics calculations.
Recent work has added many new capabilities (Figure 2), including the ability to select specific groups of atoms based on connectivity and to modify bonding patterns. Jmol can now display orbitals and other surfaces, including planes and axes that can help in the visualization of symmetry. Most recently, Jmol can now depict a wide range of polyhedral structures. These and many more capabilities will be highlighted in this paper.
Jmol supports many of the common input and output formats, and new file reading capabilities are added on an as-needed basis (Table 1). Compressed files are automatically uncompressed in the browser for faster downloads.
The Problem of Molecular
Visualization on the Web Chemistry is often referred
to as the “molecular” science. As such, students of chemistry early on are
introduced to molecules as three-dimensional objects, and many sorts of
wooden or plastic or even paper molecular models are used in the teaching of
chemistry. To this day, many students in organic chemistry are still required
to purchase molecular model “kits” that allow the construction of a limited
set of molecular structures. These kits help students “see” the symmetries
and structural factors involved in chemical geometry and reactivity. RasMol[1] was a key development in the history of
computer-based molecular visualization. Roger Sayle
began developing it in 1989 as a system for rendering 3D molecules on
raster displays. He spent several years in the early 1990s improving
performance and porting to various systems. With the help of the growing user
community, he continued to add functionality, much of it related to PDB
files and biochemistry. In 1993 he released the copyright to the public
domain and posted the source code on the internet. One of RasMol’s most
intriguing aspects was (and still is) that it has a scripting language which
can be used to control the visual representation of the molecule. One could
develop lists of operations to be carried out on the molecule in order to
showcase important aspects of the molecular structure such as alpha helixes,
beta-pleated sheets, and hydrogen bonds. RasMol is a stand-alone
program that requires downloading and installation. In the mid
1990s, with increasing interest in graphical web browsers on the internet,
there was growing demand among chemical/biochemical
educators and researchers for a method of conveying the three-dimensional
aspect of molecules on the web. Using the RasMol public domain source code
as a basis, Molecular Design Inc. created a plug-in for the Netscape
web browser. The Chime Netscape plug-in was introduced to the chemical
community at the 211th ACS National Meeting almost exactly 10 years
ago, in April of 1996.[2] Chime’s primary use was for visualizations within
MDL’s proprietary chemical inventory database system, but it was also
made freely available to the public. Chime was an enabler for
educators, providing widespread availability to a web-based scriptable
“virtual” molecular model kit along the lines of RasMol. Chime was quickly
adopted by educators as a valuable tool for displaying molecules and
demonstrating concepts. Thus, with the introduction of Chime,
virtual molecular visualization took a big step into the general chemistry,
organic and biochemistry classroom. Chime was based upon
browser plug-in technology, and therefore inherited a set of technical
complexities associated with plug-ins. A problem for end-users was that the
plug-in required manual installation on individual user machines. At the university
level in particular this was a problem, as students might be using
campus-wide public computing resources largely out of the control of the
professor or the designer of the web site. A set of larger problems
was caused by the fact that plug-in behavior was different across browsers
and operating systems. Netscape and Microsoft did not implement plug-ins
in the same way. The MSFT Internet Explorer plug-in mechanism was
different from the Netscape Navigator plug-in mechanism and Netscape on
Windows behaved differently from Netscape on Macintosh. At times this
made it difficult for content developers to build portable web pages that
would reliably run on different web browser platforms. In the long run, this
inconsistent plug-in behavior had an even more significant impact on MDL
and on the future of Chime. Porting and maintaining Chime on
different platforms represented a significant ongoing software development cost
to MDL. These costs became increasingly difficult to justify,
particularly after the internet “bubble” burst. MDL ultimately made the
decision to stop porting to new systems and to put Chime in “maintenance
mode”. Consider these notes at MDL's Chime support web site: When will you have a version
of Chime for Macintosh OS-X or Linux? We don't have any current plans
for an OS-X version of Macintosh or a Linux version. Why doesn't Chime work with
Netscape 6 and 7? Chime has been tested on a number
of platforms, including certain versions of Netscape and Internet Explorer.
The fully tested browser and operating system combinations are listed under
the Requirements link. Other combinations of operating system and browser may
work partially or not at all. Unfortunately, we don't have the resources to
test Chime on every new browser and operating system. The Jmol Solution This potential crisis for
chemical educators who were developing web-based content using Chime was
diffused by the timely development of Jmol. The Java programming
language[3] was designed as a modern programming environment
for the web. Cross-platform support and safe execution of downloaded code are
fundamental to the design. Because the Jmol applet is written in Java, most
of the cross-platform porting, maintenance and installation issues associated
with the Chime plug-in do not exist. Jmol development regularly occurs on
Linux, Mac OS X and Windows systems. When a user visits a web page containing
the Jmol applet, the program is
automatically downloaded and executed on the client’s browser without
requiring installation. Dan Gezelter started
writing Jmol in the late 1990s with the intention of building a replacement
for XMol, an early program distributed by the Egon Willighagen joined the
project in 1998, contributing support for CML files and other file formats as
well as other new functionality. In 2002 he became Jmol project leader and
oversaw a number of public releases. Egon also has responsibility for
maintaining the interface between the Chemical
Development Kit software library and Jmol. Miguel Howard joined the
project in late 2002, with the explicit goal of building Jmol into a viable
replacement for the Chime plug-in. His first major contribution was an
interpreter for the RasMol/Chime scripting language. In the spring of 2003
Miguel began designing and implementing a software-based graphics engine that
would provide cross-platform high-performance 3D functionality. He
subsequently redesigned and re-implemented the core architecture to support
efficient rendering of macromolecules with hundreds of thousands of atoms. An extended testing period began
at the end of 2003. Through 2004 a small set of users around the world made
significant contributions to the Jmol project by testing Jmol releases and
explaining scripting behaviors of RasMol and Chime. Tim Driscoll, because of
his extensive experience with Chime, was particularly helpful during this
process. He helped resolve a number of issues associated with macromolecules,
including definitions of protein/nucleic predefined sets and schematic
rendering of secondary structures. Bob Hanson also got
involved during this time, developing an interactive web site providing Jmol script documentation
and examples. Feedback and contributions from Bob and other JavaScript
experts led to the development of the Jmol.js JavaScript library, which makes
introducing Jmol into web pages almost trivial. Jmol version 10.00, a fully
functional replacement for the Chime plug-in, was released in December 2004. As the Jmol community grew,
requests for new functionality came in. Through 2005 a number of new
scripting, rendering and file format features were introduced into the
development version of Jmol. Notable additions included support for polyhedra
representations and isosurfaces. Nicolas (Nico) Vervelle
joined Jmol because of his interest in the Folding@home project. He contributed the file reader for folding@home’s files and added expressions for substructure searching using SMILES syntax. Nico then took over responsibility for managing internationalization and localization efforts. Another official release, Jmol
version 10.2, was made in April 2006. It included language localizations
for Catalan, German, Spanish, Estonian, French, Dutch, and Portuguese. In the
spirit of open source development, localizations were contributed and
maintained by Jmol users from different countries. Jmol development continues
along a variety of paths, and the Jmol community continues to grow, with
increasing involvement and interest from educators and scientists around the
world. What follows is a
discussion of the use of Jmol in education in a variety of chemical and
biochemical contexts. The discussion highlights web sites that have utilized
Jmol, general capabilities of the current version of Jmol, and a few sneak
peeks of some of the more experimental features of Jmol not yet publicly
released. This list is by no means comprehensive. Most of these examples were
taken from the extensive list at the Jmol wiki,
which also includes examples of the use of Jmol in research. Jmol for General and
Inorganic Chemistry In the area of general
chemistry, Jmol has been used primarily to illustrate the various common
three-dimensional molecular shapes. Some representative sites are shown in
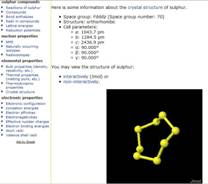
Table 2. WebElements utilizes
Jmol for the display of crystal structures of the elements. The 3Dchem.com site illustrates
how easy it is to add a structure to any web page. This can be seen by taking
a peek at the source for one of
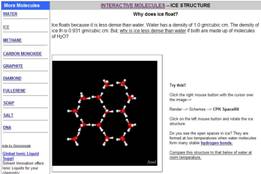
these pop-up windows. Explain
it with Molecules uses Jmol to help explain everyday questions with the
aid of 3D models. The CoolMolecules site illustrates a novel feature of Jmol –
the ability to create a 3D model of a structure on the fly – with no actual
structure file on the server. Instead, the data for all of the structures are
stored in a compressed fashion in one single
file, which is delivered to the user’s browser upon page loading. John
Gutow’s VSEPR tutorial and Mark Winter’s Introduction to
VSEPR are both excellent examples of the integration of Jmol into an
online lesson in bonding theory and its association to molecular shape and
electronic configuration. Richard
Spinney ( The issue in inorganic
chemistry for which Jmol has provided a general solution is the description
of symmetry. An example can be seen in a recent contribution at the Journal
of Chemical Education’s WebWare
site, a component of the JCE
Digital Library, which is a Collection within the National Science
Digital Library (NSDL). We
expect to see additional applications of Jmol within this area in the future
as Jmol develops and the newer features involving symmetry planes, rotational
axes, internal axis rotation, and internal coordinate referencing become more
widely known. |
|||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Jmol for Organic
Chemistry We cannot hope to present
in this paper more than a small glimpse of the extensive use of Jmol that has
appeared in the area of organic chemistry. A recent Google search of “jmol
organic” returned “about 18,400” hits. In Table 3 we give only a small
sampling the many excellent sites on the Web, just as a way of illustrating
some of the variety of uses of Jmol. Topics cover a wide range, primarily
because so much of organic chemistry relies upon a visual understanding of
molecular structure and reactivity. |
|
Table 3. A Few Representative Uses of Jmol in
Organic Chemistry |
|
|
Virtual Textbook of Organic
Chemistry
|
http://www.cem.msu.edu/~reusch/vtxtindex.htm “An interactive textbook covering the usual
topics treated in a college sophomore-level course. Links are offered to
advanced discussions of selected topics.” This understated description
accompanies the most extensive online discussion of organic chemistry ever
produced. It is, in fact, a complete, online textbook involving hundreds of
utilizations of Jmol. |
|
Dr. Spinney’s World of
Chemistry (symmetry page)
|
http://undergrad-ed.chemistry.ohio-state.edu/
Uses of Jmol at this site include the displaying of atomic orbitals based on |
|
Mol4D
|
http://cheminf.cmbi.ru.nl/wetche/organic/
“The WeTChe.NL tutorials are an initiative of the Faculty of Science, |
|
Spectral Zoo
|
http://web.centre.edu/muzyka/organic/organic.htm
“This site includes Jmol-enabled tutorials to allow students to better understand
difficult organic chemistry concepts such as conformations, sterics, and
stereochemistry. There are also Jmol-enabled animations for substitution and
elimination reactions. The Spectral Zoo is a set of combined spectra which
serve as practice problems.” |
|
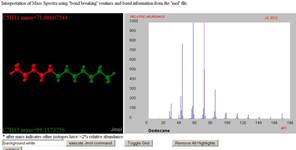
Jmol/JSpecView
|
http://wwwchem.uwimona.edu.jm/spectra/JSpecView/
msanim/dodecaneJ/dodecane.html “JSpecView is a viewer for spectral
data in the JCAMP-DX format. Examples of
the combination of spectra and molecular graphics to show the interpretation
of simple IR, MS or NMR using JspecView and Jmol
are available.” This page uses several innovative aspects of Jmol, including
“designer” planes and the use of “show file” to transmit the contents of the
model file to JavaScript for further processing – in this case, determining
the masses of the fragments selected by the user. |
|
DCU Molecular Viewing
Gallery
|
http://webpages.dcu.ie/~pratta/jmgallery/JGALLERY.HTM
“The DCU Molecular Viewing Galleries currently contain over 400 molecules,
organised alphabetically and viewable in pairs in adjacent viewing frames.” |
|
Jmol for Biochemistry
and Molecular Biology Following the precedent of Chime
and RasMol, Jmol has been utilized in the area of biochemistry and molecular
biology in a variety of applications both in research and in education.
Highlighted in Table 4 are selected applications of particular interest to
educators that are employing Jmol, including web-accessible databases (Research Collaboratory for
Structural Bioinformatics Protein Data Bank), structure explorers (FirstGlance,
JenaLib),
tutorials focusing on specific biomolecules and biological pathways (the Online Molecular Museum, Libmol,
y Apuntes
de Biología Molecular), and media tools for instructors (Molecules in Motion,
MolSlides). |
|
Table 4. Representative Uses of Jmol in Biochemistry
and Molecular Biology |
|||
|
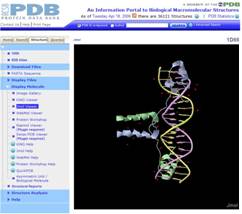
RCSB PDB (1d66 shown)
|
http://www.rcsb.org/pdb “The
RCSB PDB provides a variety of tools and resources for studying the
structures of biological macromolecules and their relationships to sequence,
function, and disease. The RCSB is a member of the wwPDB, whose mission is to
ensure that the PDB archive remains an international resource with uniform
data. This site offers tools for browsing, searching, and reporting that
utilize the data resulting from ongoing efforts to create a more consistent
and comprehensive archive.” Jmol is one of the five viewers recommended by RCSB. |
||
|
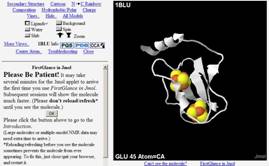
FirstGlance in Jmol (1blu
shown)
|
http://molvis.sdsc.edu/fgij
“FirstGlance in Jmol is a simple, free tool for macromolecular visualization.
The initial display is Cartoon plus Ligands+. Click on the links and buttons
above to see different aspects of the molecular structure.” Buttons show
informative views of any macromolecule available on-line. Help, including
color keys, appears automatically and is always in view. Hyperlinks show a molecule
in one click. It is designed to be useful both to novices and to specialists. |
||
|
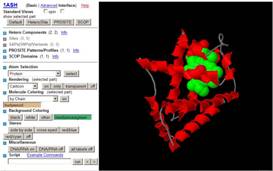
JenaLib (1ash shown)
|
http://www.imb-jena.de/IMAGE.html
“The Jena Library of Biological Macromolecules (JenaLib) is aimed at a better
dissemination of information on three-dimensional biopolymer structures with
an emphasis on visualization and analysis. It provides access to all
structure entries deposited at the Protein Data Bank (PDB) or at the Nucleic
Acid Database (NDB). In addition, basic information on the architecture of
biopolymer structures is available.” |
||
|
The Online Molecular Museum
|
http://www.callutheran.edu/Academic_Programs/ |
||
|
Libmol (1ag2 shown)
|
http://www.librairiedemolecules.education.fr Librairie de molecules “La librairie de
molécules a pour but d'améliorer le transfert des connaissances de la
recherche vers l'enseignement. Elle propose des modèles moléculaires sélectionnés
à partir des banques de données des chercheurs par des enseignants pour les
enseignants. Tout utilisateur peut contribuer à l'enrichissement de la
librairie en proposant de nouveaux modèles et de nouvelles applications
pédagogiques.” |
||
|
Apuntes de Biología Molecular
|
http://av.bmbq.uma.es/bma/apuntes/index.htm “Con esta
asignatura pretendemos enseñar las características físico-químicas de las
macromoléculas que condicionan todo el funcionamiento celular así como
mostrar que la estructura de las moléculas viene guiada por las
distribuciones electrónicas de los átomos que las componen. También esperamos
que quede claro que en la conformación y en los cambios conformacionales de
las moléculas reside su función biológica y su regulación. Con ello queremos
dejar claro que es mucho más lo que se desconoce que lo que se conoce sobre
la biología molecular, por lo que animamos a la búsqueda y al análsis crítico
de la bibliografía y los medios para acceder a la información molecular.” |
||
|
Molecules in Motion
|
http://www.moleculesinmotion.com/
This site demonstrates two
entertaining Jmol-based “movies” entitled “What is a Protein?” and “DNA Structure (with a little RNA thrown in)”. |
||
|
MolSlides (1d66 shown)
|
http://www.umass.edu/microbio/chime/ |
||
|
Jmol for Mineralogy and
Crystallography Jmol has found use in displaying
the structure of minerals. Sites utilizing Jmol are listed in Table 6. The
recently added connection and polyhedra capabilities of Jmol are expected to
expand this use considerably. |
|
Table 6. Representative Uses of Jmol in Mineralogy and
Crystallography |
|||
|
Mineralogy Structure Index
(perovskite shown)
|
http://webmineral.com/jmol/index.shtml
“This mineral database contains 4,442 individual mineral species descriptions
with links and a comprehensive image library.” Mineral Structure Index:
“Click on mineral name in the Index of Mineral Structures for a large
interactive display (Jmol Applet) of the chemical structure.” |
||
|
The Virtual Museum of
Minerals and Molecules
|
http://virtual-museum.soils.wisc.edu
“The Virtual Museum of Minerals and Molecules™ is a web-based focal point and
resource for 3-D visualizations of molecules and minerals designed for
instructional use.” This website is organized
along the lines of a typical museum with “wings” and “galleries” all devoted
to the structure of minerals. |
||
|
WebEMAPS (FeS2 shown)
|
Web Electron Microscopy
Applications Software With this website you can enter
crystal data from CIF files or manually based on symmetry or choose from a
list of crystal structures already on the server. After specifying the number
of unit cells, the structure is displayed using Jmol. |
||
|
Molecular Origami (zircon shown)
|
http://www.stolaf.edu/people/hansonr/mo Molecular Origami The recent addition of
“collapsed polyhedra” to Jmol (prototype version) has allowed the use of Jmol
to produce realistic-looking virtual equivalents of the precision scale
models created from paper. |
|
Jmol in Textbooks An exciting development has
been the utilization of Jmol by several publishers for mostly
biochemistry-related textbooks as the viewer of choice for their web site
ancillary material. Those we know of are listed in Table 5. |
|
Table 5. Textbooks Utilizing Jmol |
|
|
Fundamentals of Biochemistry: Life at the Molecular Level,
2nd Edition |
New to this edition: “Upgraded and re-designed web-based
media program to accompany and complement the text includes Interactive
Exercises, Guided Explorations, Animated Figures, as well as Kinemages. Media
resources have been upgraded to take advantage of the most recent,
web-friendly 3D visualization tools, eliminating the need for the Chime
plug-in, which has proven to be problematic.” |
|
“The Second Edition has
been thoroughly revised and updated to incorporate the latest scientific
findings on popular topics such as disease-causing organisms and genetic
defects. Case study chapters have been placed throughout the book to tie
real-life scenarios into the concepts that follow. Two of the book’s key
pedagogical features, Discovery Questions and Math Minutes, have also been
updated and expanded. The interactive companion website has been reprogrammed
with JMOL, the latest 3-D software used to view DNA structures.” |
|
|
Brooks-Cole General,
Organic, and Biochemistry Chemistry Textbooks |
New to this edition: “Organic and Biochemistry OWL
content takes advantage of the latest technological advances in online computer
modeling using Jmol and Marvin Sketch. MarvinSketch, a Java applet for
viewing and drawing chemical structures, enables OWL to grade chemical
structures that the students draw and is used extensively in the content for
the Organic and Biochemistry chapters. Jmol, an interactive molecule viewer,
enables students to rotate molecules, to change the display mode (ball and
stick, space fill, etc.), and to measure bond distances and angles.” |
|
Lehninger: Principles of
Biochemistry |
Future edition will incorporate
Jmol. |
|
Stryer: Biochemistry 6/e |
Future edition will
incorporate Jmol. |
|
Jmol in the Hands of
Instructors and Students Examples of individual
instructors involving students in the use of Jmol are starting to appear on
the Web. For example, the online homework system, webassign, has recently
added a Jmol template. Jmol has also been integrated into the OWL system. Students at the |
|
Conclusions -- Future
Directions for Jmol Jmol development relies solely
on the volunteer contributions of scientists, educators, and programmers.
Nonetheless, it is expected to be rapid during 2006. Areas currently under
active development are listed below. Links open experimental pages that
illustrate implementation of these goals within the very latest prototype
“trunk” of the development tree (trunk). We
welcome your comments, and we encourage you to get involved by joining the jmol users
list if you are interested in contributing to the discussion or the Jmol developers list
if you are interested in contributing at the developer level. [after-notes added 2/23/2007] ·
The
introduction of solvent-accessible
surfaces and surface mapping.
[see isosurface]
·
The
ability to apply crystallographic
symmetry and view multiple unit cells.
[see load]
·
The
calculation and display of dipole
moments and bond dipoles.
[see dipole]
·
Drawing points, lines, and
planes based on atom positions or {x y z} coordinates.
[see draw]
·
Rotation and spinning around
internal axes.
[see rotate]
·
Making
multiple measurements all at once and providing running measurements in
animations.
[see measure]
·
The
ability to extract calculational
and structural data from files.
[see mo]
·
The
ability to probe the applet
for a wide range of status messages as a replacement for callback functions.
[callbacks found to be superior to polling -- see callback]
·
Easier
integration into wikis such as MediaWiki. ·
Support
for multiple file loading and independent manipulation.
[see invertSelected]
·
Further
development of the application programming interface (API), which allows Jmol
to “talk” to other applications and applets.
[see AJAX]
Clearly, Jmol has come a long way during the past few years and is rapidly becoming one of the most popular Java applets in chemistry. Its strength is in its speed of rendering, its flexibility in file reading, its broad applicability in both small-molecule and macromolecule applications, and its ease of use. Applications range from general chemistry to computational chemistry, from inorganic chemistry to molecular biology to mineralogy and crystallography. The applet is fast becoming far more than simply a tool for putting a “gee-wiz” three-dimensional model of a molecule on the web. New capabilities allow it to be an investigative tool as well, allowing the web designer/user to gain access to information such as atomic charges, dipoles, vibrational frequencies, space group symmetries, and molecular orbitals contained in data-rich formats such as CIF, CDX, CML, and SMOL. Most importantly, as an open-source project, the Jmol discussion lists have become valuable forums for the discussion of important molecular visualization and data analysis ideas among an energetic and dedicated group of scientists and educators from all over the world. |
References