
Jmol._Canvas2D (Jmol) "jmol1"[x]
loading... j2s/core/package.js
loading... j2s/core/core.z.js -- required by ClazzNode
Jmol JavaScript applet jmol1_object__246769292143953__ initializing
Jmol getValue debug null
Jmol getValue logLevel null
Jmol getValue allowjavascript null
AppletRegistry.checkIn(jmol1_object__246769292143953__)
viewerOptions:
{ "name":"jmol1_object","applet":true,"documentBase":"https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/cover.htm","platform":"J.awtjs2d.Platform","fullName":"jmol1_object__246769292143953__","codePath":"https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/j2s/","display":"jmol1_canvas2d","signedApplet":"true","appletReadyCallback":"Jmol._readyCallback","statusListener":"[J.appletjs.Jmol object]","syncId":"246769292143953","bgcolor":"white" }
setting document base to "https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/cover.htm"
(C) 2012 Jmol Development
Jmol Version: 14.1.11_2014.03.07 2014-03-07 14:38:
java.vendor: Java2Script (HTML5)
java.version: JSmol 14.1.11 Mar 1, 2014
os.name: Mozilla/5.0 AppleWebKit/537.36 (KHTML, like Gecko; compatible; ClaudeBot/1.0; +claudebot@anthropic.com)
Access: ALL
memory: 0.0/0.0
processors available: 1
useCommandThread: false
appletId:jmol1_object (signed)
Jmol getValue emulate null
defaults = "Jmol"
Jmol getValue boxbgcolor null
Jmol getValue bgcolor white
backgroundColor = "white"
Jmol getValue ANIMFRAMECallback null
Jmol getValue APPLETREADYCallback Jmol._readyCallback
APPLETREADYCallback = "Jmol._readyCallback"
Jmol getValue ATOMMOVEDCallback null
Jmol getValue CLICKCallback null
Jmol getValue ECHOCallback null
Jmol getValue ERRORCallback null
Jmol getValue EVALCallback null
Jmol getValue HOVERCallback null
Jmol getValue LOADSTRUCTCallback null
Jmol getValue MEASURECallback null
Jmol getValue MESSAGECallback null
Jmol getValue MINIMIZATIONCallback null
Jmol getValue PICKCallback null
Jmol getValue RESIZECallback null
Jmol getValue SCRIPTCallback null
Jmol getValue SYNCCallback null
Jmol getValue STRUCTUREMODIFIEDCallback null
Jmol getValue doTranslate null
language=en_US
Jmol getValue popupMenu null
Jmol getValue script null
Jmol getValue loadInline null
Jmol getValue load null
Jmol applet jmol1_object__246769292143953__ ready
loading... j2s/core/corescript.z.js
script 1 started
loading... j2s/core/corezip.z.js
FileManager opening 1 https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png
FileManager checking PNGJ cache for https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|JmolManifest.txt
ZipUtil cached 59230 bytes from https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png
https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|state.spt
https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|JmolManifest.txt
https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|1hxw.pdb.gz
https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png
https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|Jmol_version_13.0.7_dev__2012-10-12_22.06
FileManager opening 3 https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|JmolManifest.txt
FileManager checking PNGJ cache for https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|JmolManifest.txt
FileManager using memory cache https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|JmolManifest.txt
FileManager checking PNGJ cache for https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|state.spt
FileManager using memory cache https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|state.spt
loading... j2s/JV/StateCreator.js
loading... j2s/JV/JmolStateCreator.js -- required by JV.StateCreator
FileManager.getAtomSetCollectionFromFile(https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|1hxw.pdb.gz)
FileManager checking PNGJ cache for https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|1hxw.pdb.gz
FileManager using memory cache https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|1hxw.pdb.gz
The Resolver thinks Pdb
loading... j2s/core/corebio.z.js
loading... j2s/J/render/MeshRenderer.js -- required by J.renderbio.BioShapeRenderer
loading... j2s/J/shape/Mesh.js
loading... j2s/JW/Hermite.js
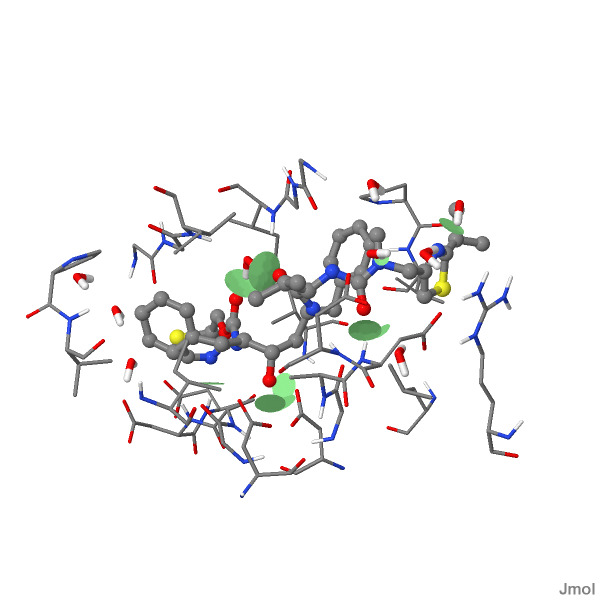
HYDROLASE/HYDROLASE INHIBITOR 24-JAN-97 1HXW
HIV-1 PROTEASE DIMER COMPLEXED WITH A-84538
loading... j2s/core/coresym.z.js
found biomolecule 1: A, B
biomolecule 1: number of transforms: 1
Time for openFile(https://chemapps.stolaf.edu/jmol/jsmol-2014-03-08/data/1hxw.png|1hxw.pdb.gz): 1581 ms
reading 2202 atoms
ModelSet: haveSymmetry:false haveUnitcells:true haveFractionalCoord:false
1 model in this collection. Use getProperty "modelInfo" or getProperty "auxiliaryInfo" to inspect them.
Default Van der Waals type for model set to Babel
2202 atoms created
ModelSet: autobonding; use autobond=false to not generate bonds automatically
loading... j2s/core/corescriptmath.z.js
unrecognized SET parameter in Jmol state script (set anyway): dynamicmeasurements
loading... j2s/core/coretext.z.js
loading... j2s/core/corescriptcmd.z.js
loading... j2s/J/shapesurface/Contact.js
loading... j2s/core/coresurface.z.js -- required by J.shapesurface.Contact
loading... j2s/J/shape/MeshCollection.js -- required by J.shapesurface.Isosurface
loading... j2s/JW/TriangleData.js -- required by J.jvxl.calc.MarchingCubes
loading... j2s/J/api/Triangulator.js -- required by JW.TriangleData
loading... j2s/J/constant/EnumHBondType.js
loading... j2s/JW/ContactPair.js
contact ID "contact1" ({1852:1901}) hbond;
Contact pairs: 17
loading... j2s/J/jvxl/readers/IsoIntersectReader.js
10 contacts with net volume 5.004599623945453 A^3
contact ID "contact1" fill noMesh noDots notFrontOnly fullylit;
loading... j2s/J/shape/Bbcage.js
loading... j2s/J/shape/Uccage.js
1938 atoms hidden
loading... j2s/J/rendersurface/ContactRenderer.js
loading... j2s/J/render/BbcageRenderer.js
loading... j2s/J/render/CageRenderer.js -- required by J.render.BbcageRenderer
loading... j2s/J/render/UccageRenderer.js