loadScript ../../jsmol/j2s/core/package.js
loadScript ../../jsmol/j2s/core/corejmol.z.js
loadScript ../../jsmol/j2s/J/translation/PO.js
loadScript ../../jsmol/j2s/core/corescript.z.js
JSmol exec jmolApplet0 start applet null
Jmol JavaScript applet jmolApplet0__214711058716917__ initializing
Jmol getValue debug null
Jmol getValue logLevel null
Jmol getValue allowjavascript null
AppletRegistry.checkIn(jmolApplet0__214711058716917__)
vwrOptions:
{ "useCommandThread":"false","animFrameCallback":"animFrameCallback","appletReadyCallback":"Jmol._readyCallback","atomMovedCallback":"atomMovedCallback","errorCallback":"errorCallback","hoverCallback":"hoverCallback","loadStructCallback":"loadStructCallback","messageCallback":"messageCallback","measureCallback":"measureCallback","pickCallback":"pickCallback","scriptCallback":"scriptCallback","applet":true,"name":"jmolApplet0","syncId":"214711058716917","bgcolor":"#000033","signedApplet":"true","platform":"J.awtjs2d.Platform","display":"jmolApplet0_canvas2d","documentBase":"https://chemapps.stolaf.edu/jmol/docs/examples-12/new.htm?topic=95","codePath":"https://chemapps.stolaf.edu/jmol/docs/examples-12/../../jsmol/j2s/","fullName":"jmolApplet0__214711058716917__","statusListener":"[J.appletjs.Jmol object]" }
setting document base to "https://chemapps.stolaf.edu/jmol/docs/examples-12/new.htm?topic=95"
(C) 2015 Jmol Development
Jmol Version: 16.2.31 2024-09-14 12:17
java.vendor: Java2Script (HTML5)
java.version: 2022-06-24 05:54:49 (JSmol/j2s)
os.name: Mozilla/5.0 AppleWebKit/537.36 (KHTML, like Gecko; compatible; ClaudeBot/1.0; +claudebot@anthropic.com)
Access: ALL
memory: 0.0/0.0
processors available: 1
useCommandThread: false
appletId:jmolApplet0 (signed)
Jmol getValue emulate null
defaults = "Jmol"
Jmol getValue boxbgcolor null
Jmol getValue bgcolor #000033
backgroundColor = "#000033"
Jmol getValue ANIMFRAMECallback animFrameCallback
ANIMFRAMECallback = "animFrameCallback"
StatusManager ANIMFRAMEcallback set f=animFrameCallback
Jmol getValue APPLETREADYCallback Jmol._readyCallback
APPLETREADYCallback = "Jmol._readyCallback"
StatusManager APPLETREADYcallback set f=Jmol._readyCallback
Jmol getValue ATOMMOVEDCallback atomMovedCallback
ATOMMOVEDCallback = "atomMovedCallback"
StatusManager ATOMMOVEDcallback set f=atomMovedCallback
Jmol getValue AUDIOCallback null
Jmol getValue CLICKCallback null
Jmol getValue DRAGDROPCallback null
Jmol getValue ECHOCallback null
Jmol getValue ERRORCallback errorCallback
ERRORCallback = "errorCallback"
StatusManager ERRORcallback set f=errorCallback
Jmol getValue EVALCallback null
Jmol getValue HOVERCallback hoverCallback
HOVERCallback = "hoverCallback"
StatusManager HOVERcallback set f=hoverCallback
Jmol getValue IMAGECallback null
Jmol getValue LOADSTRUCTCallback loadStructCallback
LOADSTRUCTCallback = "loadStructCallback"
StatusManager LOADSTRUCTcallback set f=loadStructCallback
Jmol getValue MEASURECallback measureCallback
MEASURECallback = "measureCallback"
StatusManager MEASUREcallback set f=measureCallback
Jmol getValue MESSAGECallback messageCallback
MESSAGECallback = "messageCallback"
StatusManager MESSAGEcallback set f=messageCallback
Jmol getValue MINIMIZATIONCallback null
Jmol getValue MODELKITCallback null
Jmol getValue PICKCallback pickCallback
PICKCallback = "pickCallback"
StatusManager PICKcallback set f=pickCallback
Jmol getValue RESIZECallback null
Jmol getValue SCRIPTCallback scriptCallback
SCRIPTCallback = "scriptCallback"
StatusManager SCRIPTcallback set f=scriptCallback
Jmol getValue SELECTCallback null
Jmol getValue SERVICECallback null
Jmol getValue STRUCTUREMODIFIEDCallback null
Jmol getValue SYNCCallback null
Jmol getValue doTranslate null
language=en_US
Jmol getValue popupMenu null
Jmol getValue script null
Jmol getValue loadInline null
Jmol getValue load null
Jmol applet jmolApplet0__214711058716917__ ready
script 1 started
defaultDirectory = "data"
FileManager.getAtomSetCollectionFromFile(https://chemapps.stolaf.edu/jmol/docs/examples-12/data/caffeine.mol)
callback ANIMFRAME failed TypeError: Cannot read properties of null (reading 'apply')
callback ANIMFRAME failed TypeError: Cannot read properties of null (reading 'apply')
FileManager opening url https://chemapps.stolaf.edu/jmol/docs/examples-12/data/caffeine.mol
The Resolver thinks Mol
C8H10N4O2
APtclcactv10291122543D 0 0.00000 0.00000
C8H10N4O2
Time for openFile(https://chemapps.stolaf.edu/jmol/docs/examples-12/data/caffeine.mol): 441 ms
reading 24 atoms
ModelSet: haveSymmetry:false haveUnitcells:false haveFractionalCoord:false
1 model in this collection. Use getProperty "modelInfo" or getProperty "auxiliaryInfo" to inspect them.
Default Van der Waals type for model set to Babel
24 atoms created
ModelSet: not autobonding; use forceAutobond=true to force automatic bond creation
callback ANIMFRAME failed TypeError: Cannot read properties of null (reading 'apply')
callback LOADSTRUCT failed TypeError: Cannot read properties of null (reading 'apply')
Time for creating model: 43 ms
C8H10N4O2
antialiasDisplay = true
Script completed
Jmol script terminated
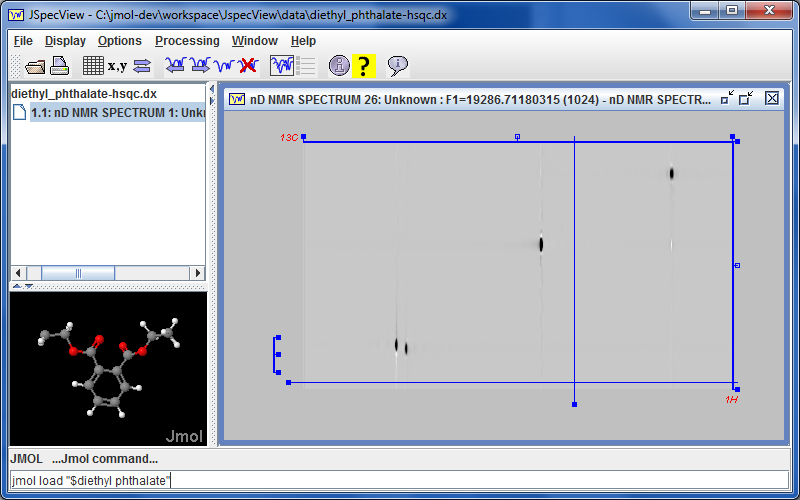 marks a new collaboration with Robert Lancashire and the
marks a new collaboration with Robert Lancashire and the  The Jmol 12.3.10 application animation play forward/reverse buttons now allow continuous animation by holding the button down.
The Jmol 12.3.10 application animation play forward/reverse buttons now allow continuous animation by holding the button down.
